Welcome to LCRAnnotationsDB
This database is a collection of information about low-complexity regions (LCRs) obtained from different sources including popular databases and methods. Here we describe the most important parts of the LCRAnnotationsDB interface.
Identification of LCRs
We used the SEG method with different parameters to identify LCRs in the UniProtKB/Swiss-Prot database:
- strict SEG (W=15, K2(1)=1.5, K2(2)=1.8)
- default SEG (W=12, K2(1)=2.2, K2(2)=2.5)
where:
- W - window size
- K2(1) - trigger complexity
- K2(2) - extension complexity
Database access
The current version of LCRAnnotationsDB contains LCRs obtained from the UniProtKB/Swiss-Prot database (version: 2023_05). The other databases used to annotate LCRs are:
- ELM (version: 1.4),
- DisProt (version: 2023_12),
- NCBI RefSeq (version: 220),
- TOPDOM (version: 3.0),
- InterPro (version: 96.0),
- RCSB PDB (API search by sequence with mmseq2 default parameters, access date 23.12.2023).
- UniProtKB (version: 2023_05),
- neXtProt (version: 2023-09-11).
- PhaSePro (version: 1.1).
- PhaSepDB (version: 2.1).
Other methods
In order to enlarge the annotations, we used three methods to identify domains, cytoplasmic regions and disordered regions that are composed of LCRs:
- IUPred3 identifies short and long disorder regions, where this database store only long regions.
- Phobius predicts cytoplasmic and non-cytoplasmic regions in proteins. We used default parameters.
Stored data
These tables and columns represent the most important elements of the LCRAnnotationsDB database.
Searching in LCRAnnotationsDB
LCRAnnotationsDB contains two main parts which can be used to search in: search form (Home Page) and data table (Database table).
Home page
The homepage allows a user to search for records using UniProt ACC, annotations, categories, or Gene Ontology categories. To select the type of data, the user should change the search option and adjust the remaining parameters to suit the data and expected results. The parameters and the outcomes of their changes are described in this chapter.
Search options
The search form allows an user to search for records using UniProt ACC, annotations, categories, or Gene Ontology categories. Input data type can be selected in dropdown list. The example button allows to fill text box with example input data, that were also used in use case chapter of publication.
For details about example data see the chapter "Use Case" in article XXXX.
Results table format
We have also added the ability to select the format of the table that will be shown as the search result. The following shows sample results of the database using different table output formats. Each contains elements common to UniProt ACC and unique to itself, among others.
Result table with low complexity regions
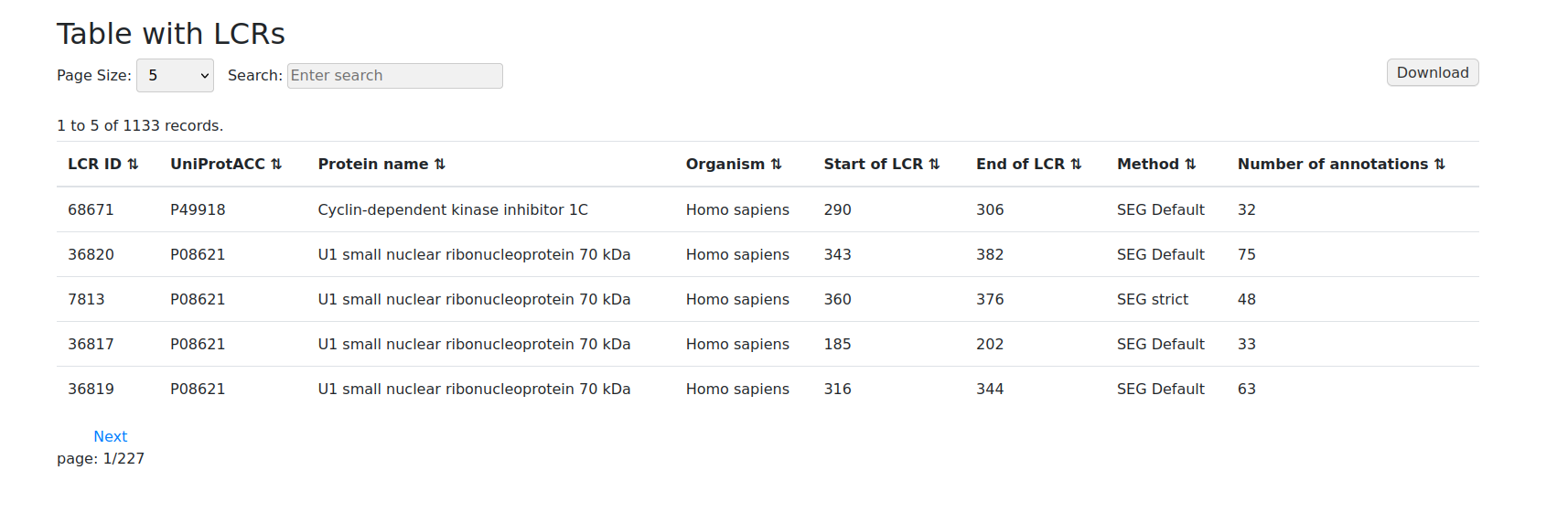
Low complexity regions table format ranks search results based on a unique LCR ID. These can also be pulled out from information about the UniProt ACC, the name of the protein, the organism and data.
Result table with proteins
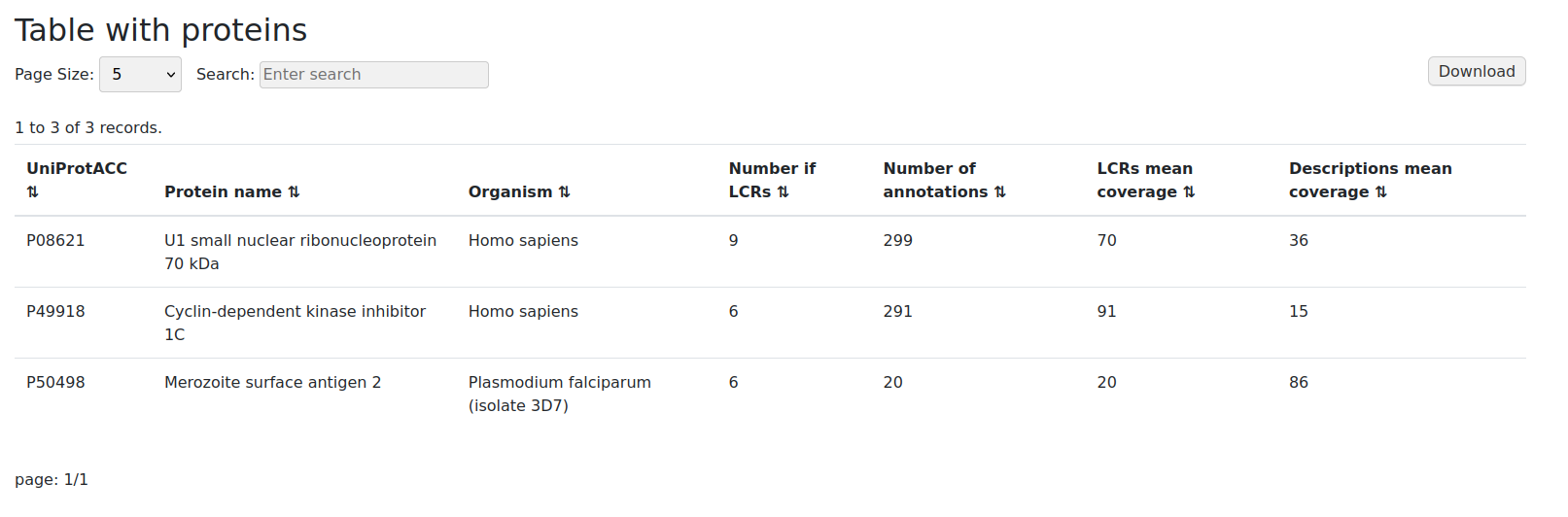
Protein table format will provide user with the information about the protein(s) of interest only. Data presented shows how many LCRs were found in each protein and the number of annotations assigned to them.
Result table with annotations
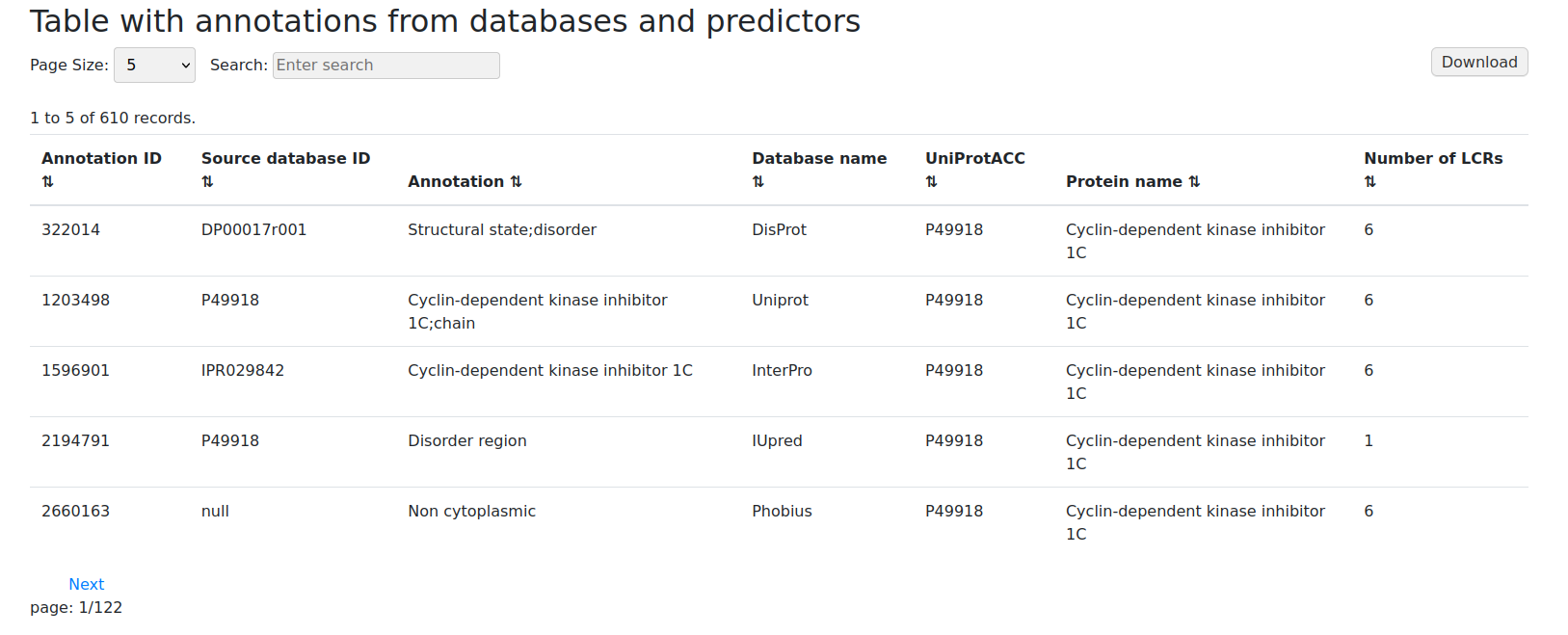
For users mainly interested in the functionality of regions, we recommend choosing annotations table format. It is sorted by the unique Annotation ID. In addition, we can preview what functional annotation is assigned and check its source (source database ID and the name of the database from which the annotation comes). User can also see UniProt ACC for easier analyses including functional comparisons of similar domains from different proteins.
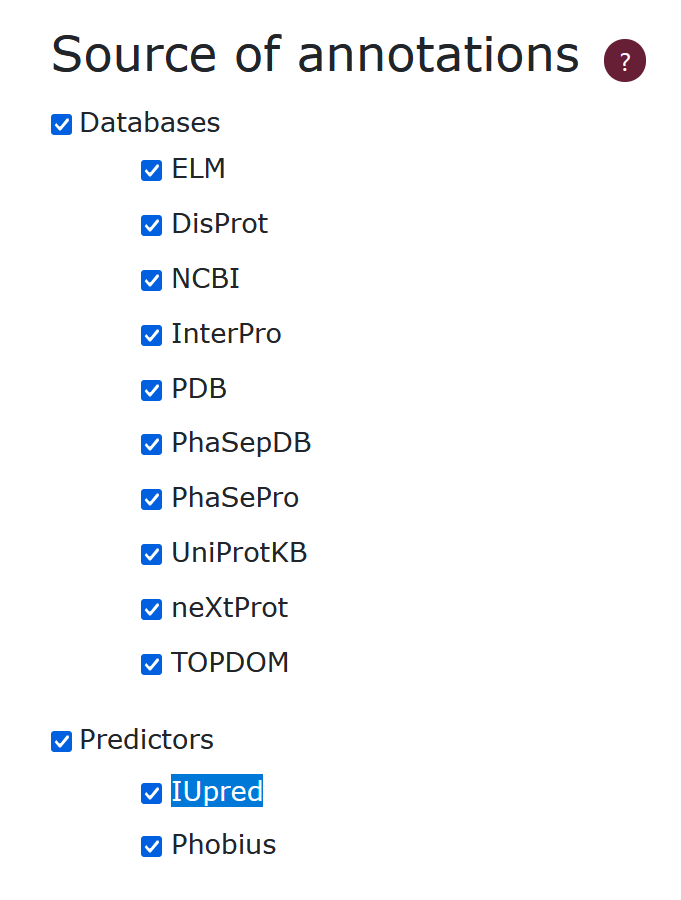
Source of data
Our database includes data from various sources. We have acquired data from 10 databases (ELM, DisProt, NCBI, InterPro, PDB, PhaSepDB, PhaSePro, UniProtKB, neXtProt, TOPDOM) and 2 methods (IUpred, Phobius). To exclude data from a certain source, the user can unmark it. A detailed description of the sources is available in the supplementary file of the publication.
Coverage
An additional option is to set sequence and/or annotation coverage. Two options can be chosen - each in the range of 0-100, where 0 means disabling this option and 100 means full coverage.
Depending on the needs, an user has a choice:
- Coverage of annotations by LCRs - calculated as 100%* (common_part_of_annotation_and_lcr)/(len_of_annotation),
- Coverage of LCRs by annotations - calculated as 100%* (common_part_of_annotation_and_lcr)/(len_of_lcr).
Both options can be left off (the default option).
Based on regions from Figure 7, coverage of annotation 1 by LCR 1 is equal to 100%*8/12=66,7% and coverage of LCR 1 by annotation 1 is 100%*8/10=80%. Analogous, coverage of annotation 2 by LCR 2 is equal to 100%*16/66=24,2% and coverage of LCR 2 by annotation 2 is 100%.
Database page
In the database subpage, user can screen database records. For each type of data, there are different subpage types (see figure 8). For example in the LCR dataset, there are only subpages with annotations of selected LCRs. In order to see all annotations from proteins, user has to select a protein dataset.
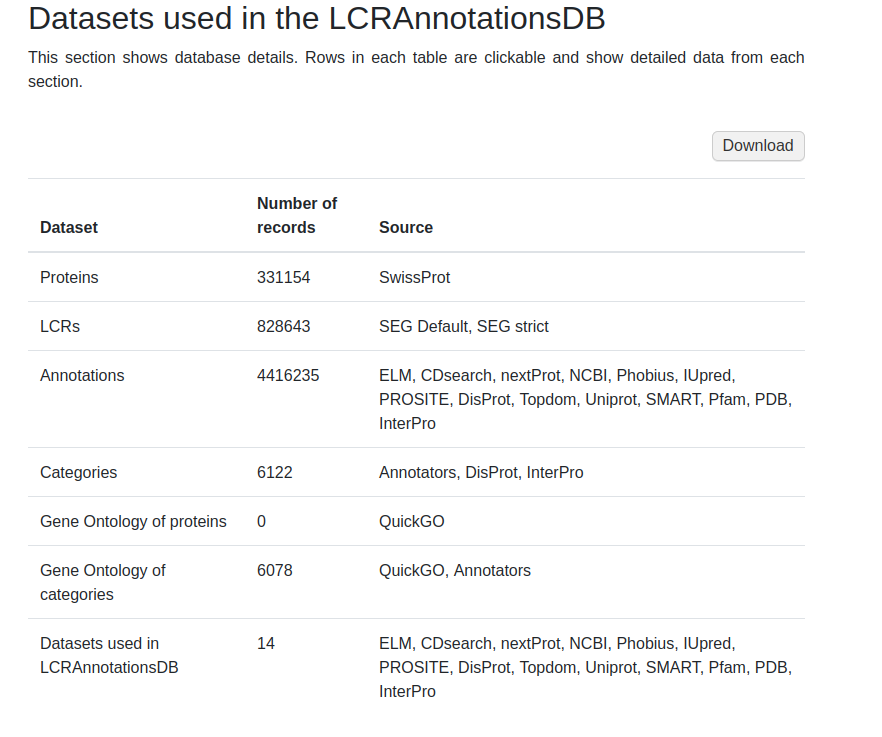
Below is a more detailed description of each subpage in datasets (see LCR, protein, category and annotation subpages) to help users to pull out the information they need in the most accessible way.
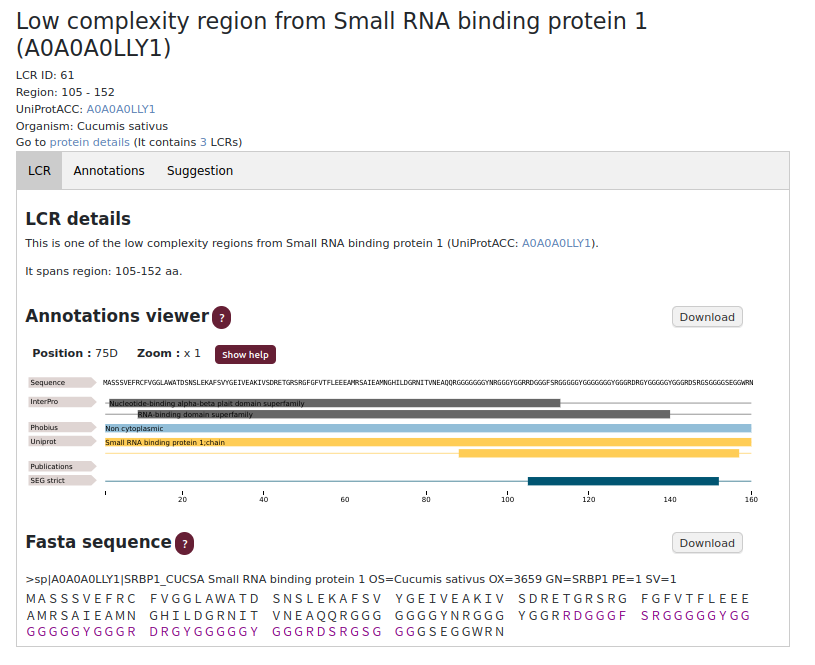
LCR subpage
The LCR subpage allows user to analyse one selected region. Information is provided about its location (beginning and end) and the location in the FASTA sequence. Annotations viewer shows the sources the LCR information was found in and what method was used for identification (SEG/SEG strict or both). In this subpage there are only annotations assigned to a selected LCR. This subpage links to the protein subpage (Go to protein details).
Protein subpage
The Protein subpage presentsthe number of LCRs in a selected protein. By moving the cursor over the Annotations viewer, user can see information about the selected LCR. User can also navigate to all annotations assigned to the LCRs in a protein. The FASTA sequence has highlighted all of the low complexity regions present in the selected protein.
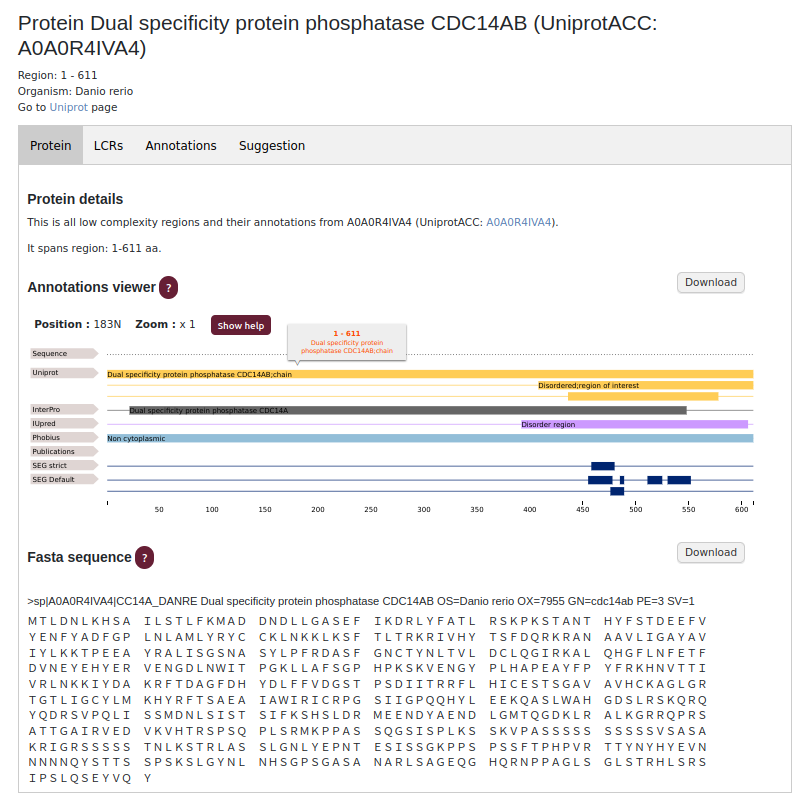
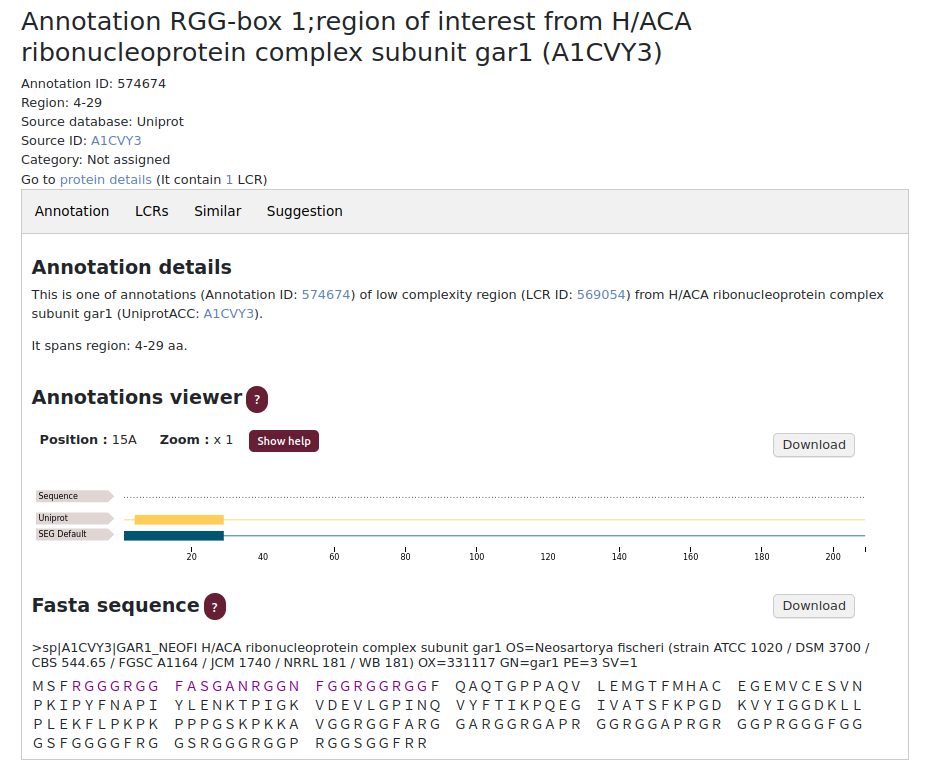
Annotation subpage
The annotation subpage contains basic information about the assigned LCRs and functional information such as: the function (category), the source of the information and its reference number from the source database (if it exists). As in the previous described subpage, the Annotations viewer and FASTA sequence are available for easier analysis/understanding of the results.
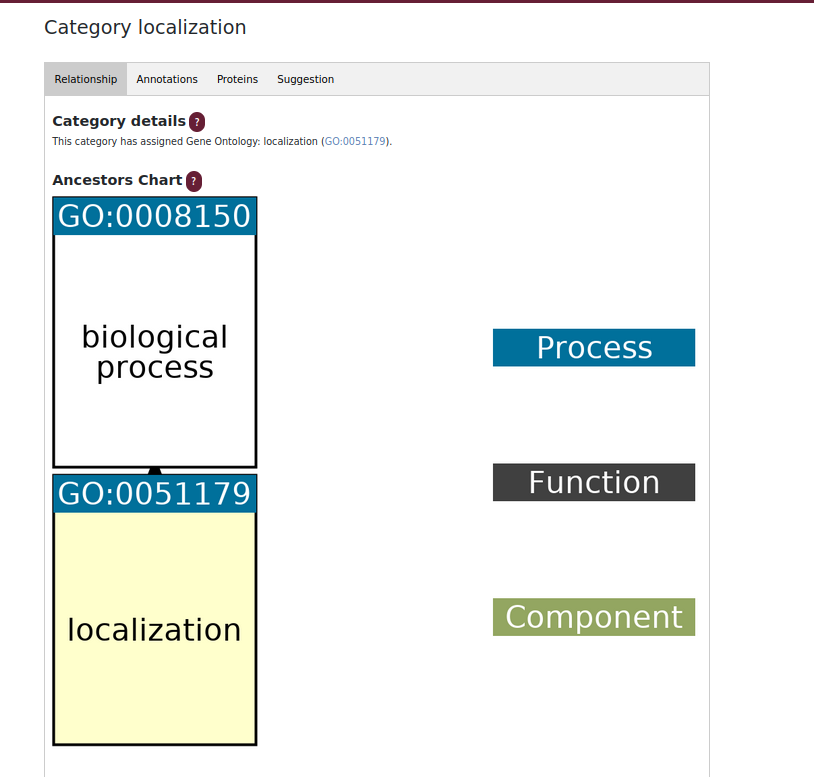
Category subpage
The category page is based on information from Gene Ontology. Ancestors chart allows user to analyse processes and functions and their relationships. Each annotation has a GO ID associated with it, so after a click user is redirected to the Gene Ontology database page.